Map reduce with examples
MapReduce
Problem: Can’t use a single computer to process the data (take too long to process data).
Solution: Use a group of interconnected computers (processor, and memory independent).
Problem: Conventional algorithms are not designed around memory independence.
Solution: MapReduce
Definition. MapReduce is a programming paradigm model of using parallel, distributed algorithims to process or generate data sets. MapRedeuce is composed of two main functions:
Map(k,v): Filters and sorts data.
Reduce(k,v): Aggregates data according to keys (k).
MapReduce Phases
MapReduce is broken down into several steps:
- Record Reader
- Map
- Combiner (Optional)
- Partitioner
- Shuffle and Sort
- Reduce
- Output Format
Record Reader
Record Reader splits input into fixed-size pieces for each mapper.
- The key is positional information (the number of bytes from start of file) and the value is the chunk of data composing a single record.
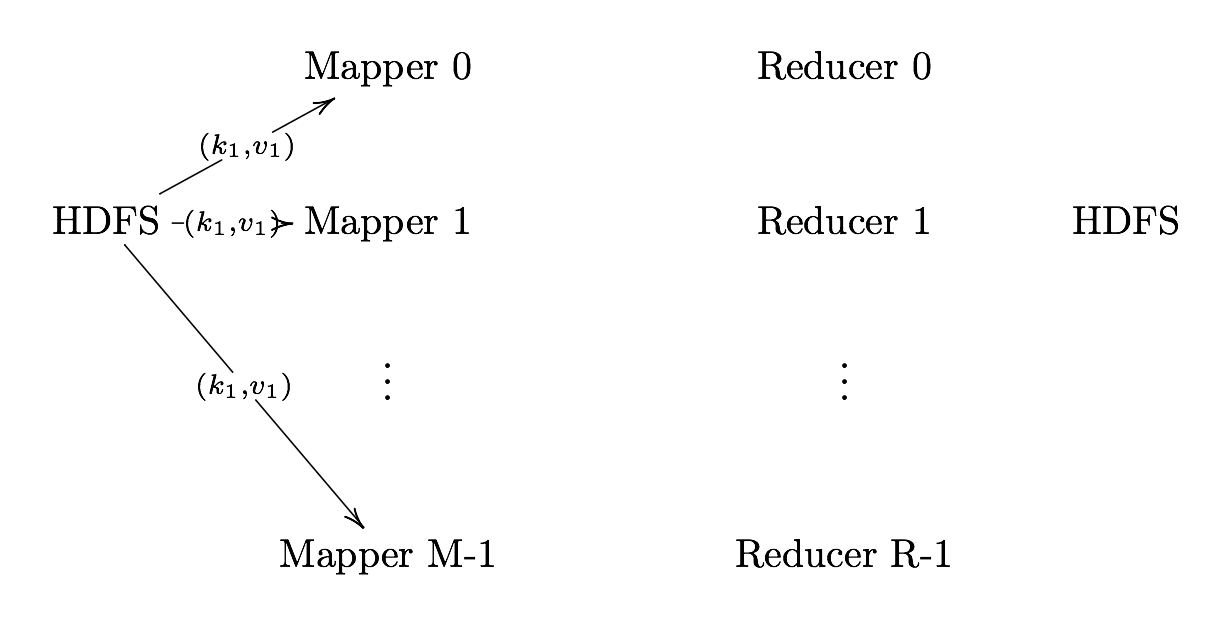
- In hadoop, each map task’s is an input split which is usually simply a HDFS block
- Hadoop tries scheduling map tasks on nodes where that block is stored (data locality)
- If a file is broken mid-record in a block, hadoop requests the additional information from the next block in the series
Map
Map User defined function outputing intermediate key-value pairs
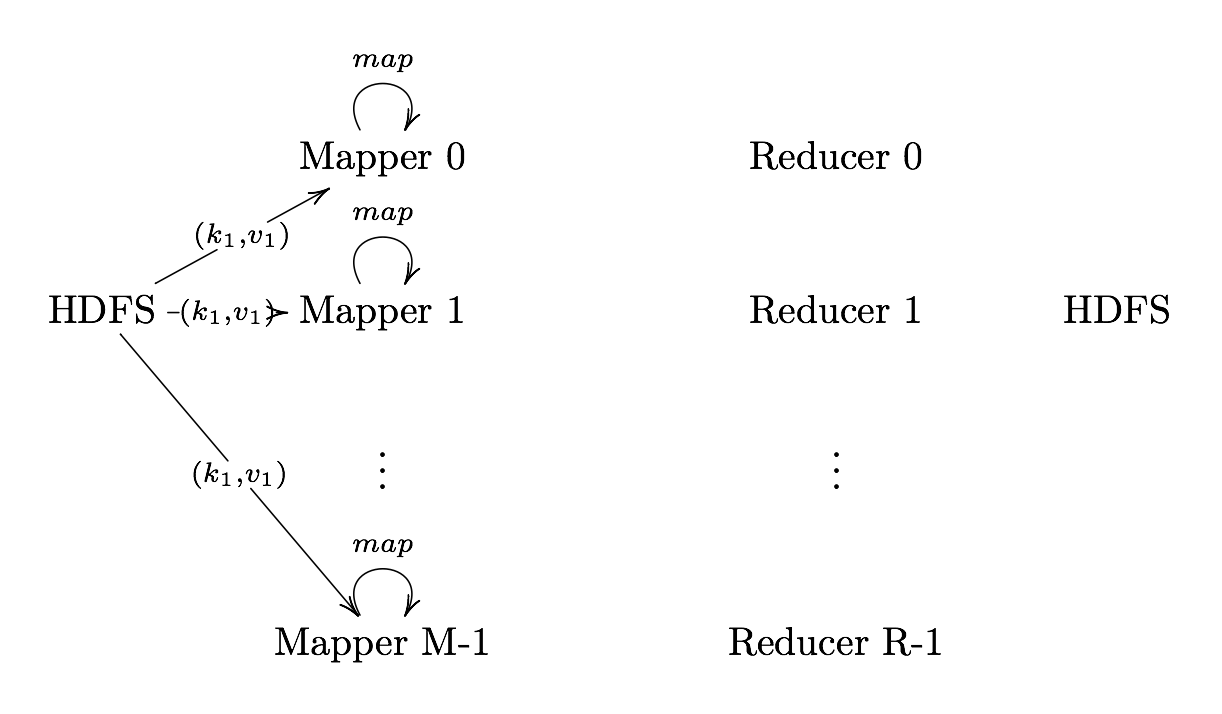
key (): Later, MapReduce will group and possibly aggregate data according to these keys, choosing the right keys is here is important for a good MapReduce job.
value (): The data to be grouped according to it’s keys.
Combiner (Optional)
Combiner UDF that aggregates data according to intermediate keys on a mapper node
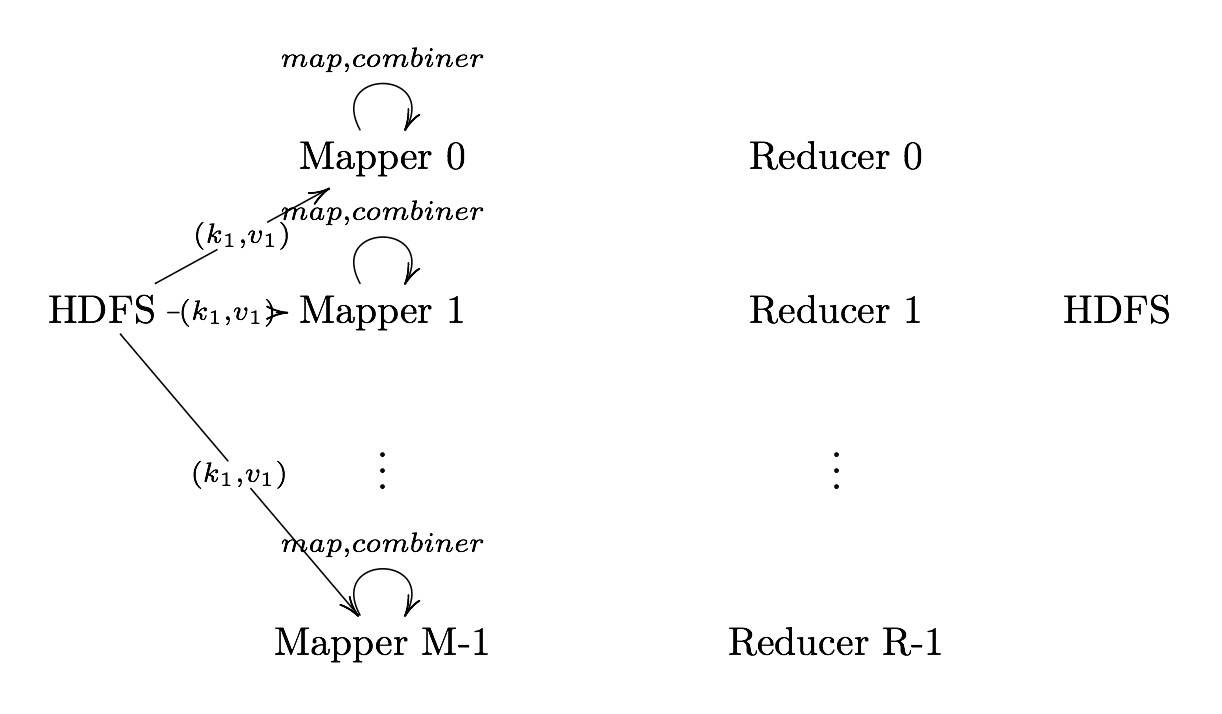
- This can usually reduce the amount of data to be sent over the network increasing efficiency
-
Combiner should be written with the idea that it is executed over most but not all map tasks. ie.
-
Usually very similar or the same code as the reduce method.
Partitioner
Partitioner Sends intermediate key-value pairs (k,v) to reducer by
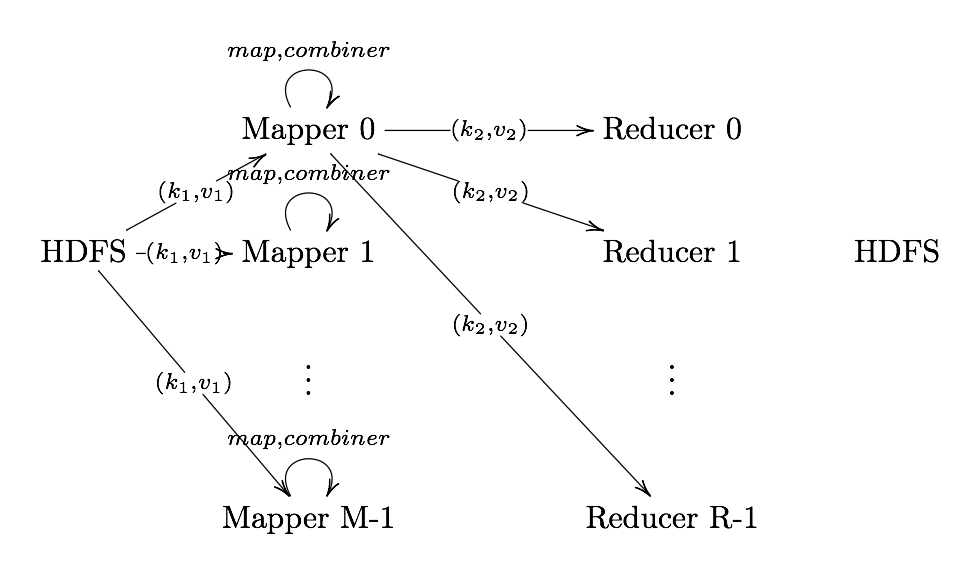
- will usually result in a roughly balanced load accross the reducers while ensuring that all key-value pairs are grouped by their key on a single reducer.
- A balancer system is in place for the cases when the key-values are too unevenly distributed.
- In hadoop, the intermediate keys () are written to the local harddrive and grouped by which reduce they will be sent to and their key.
Shuffle and Sort
Shuffle and Sort On reducer node, sorts by key to help group equivalent keys
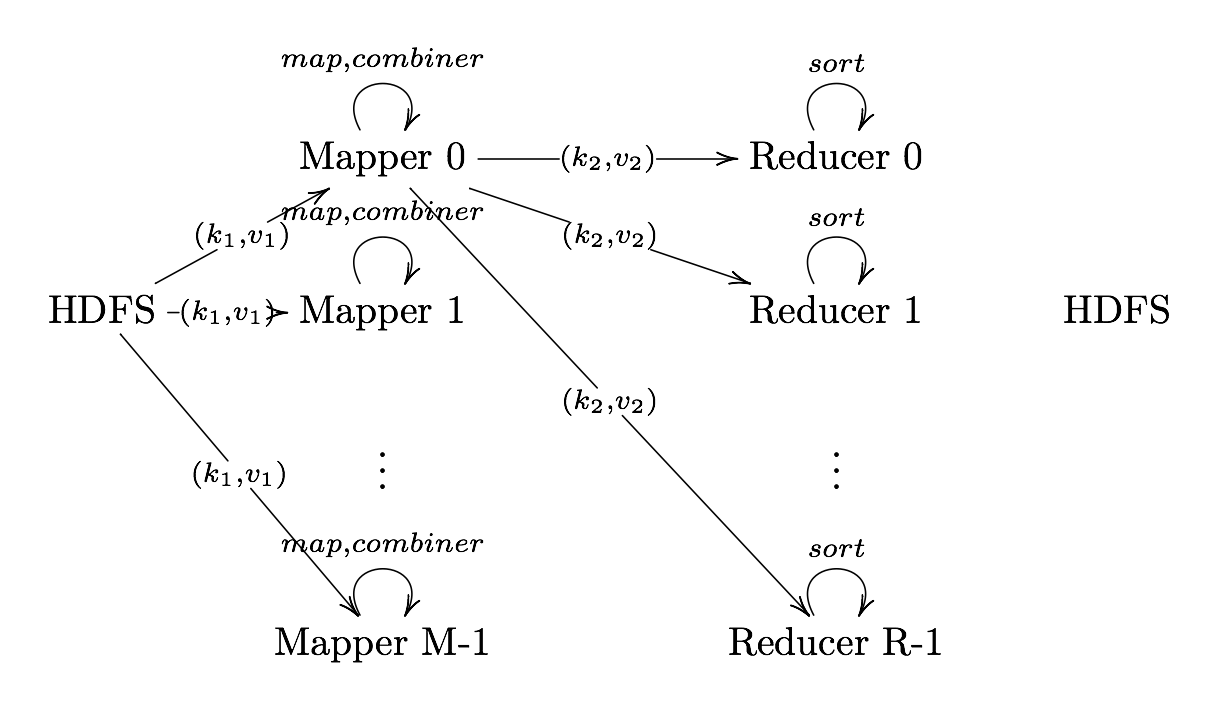
Reduce
Reduce User Defined Function that aggregates data (v) according to keys (k) to send key-value pairs to output
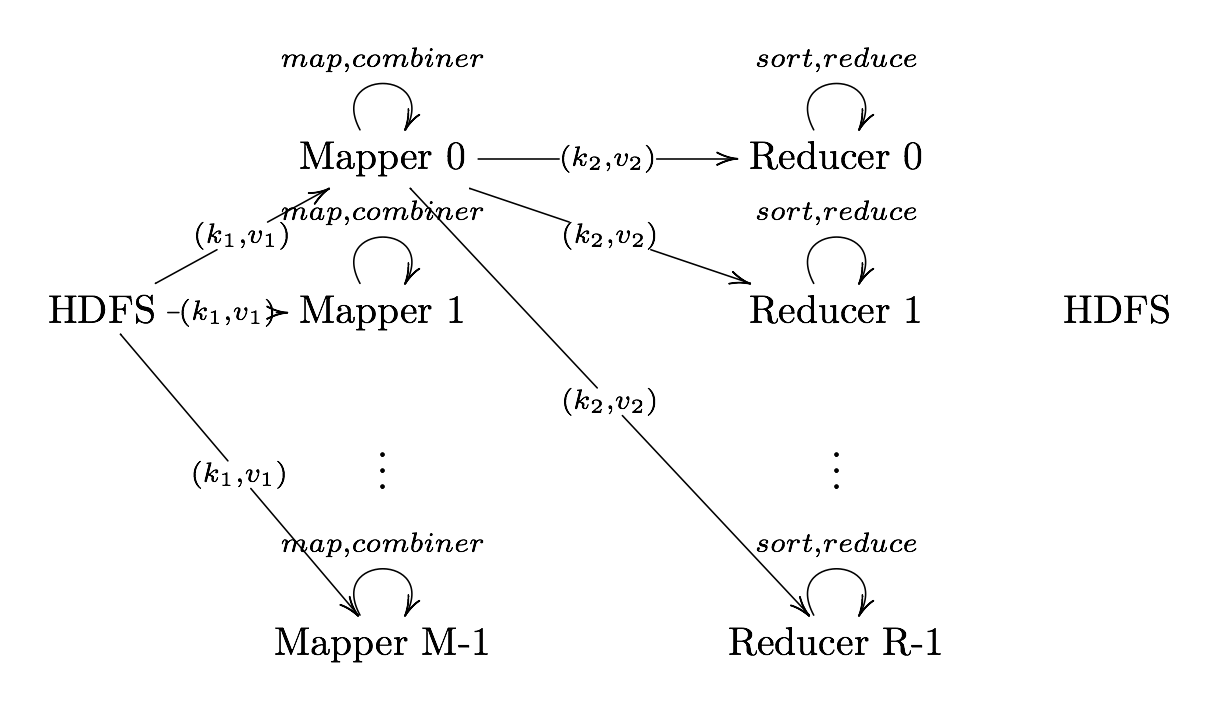
Output Format
Output Format Translates final key-value pairs to file format (tab-seperated by default).
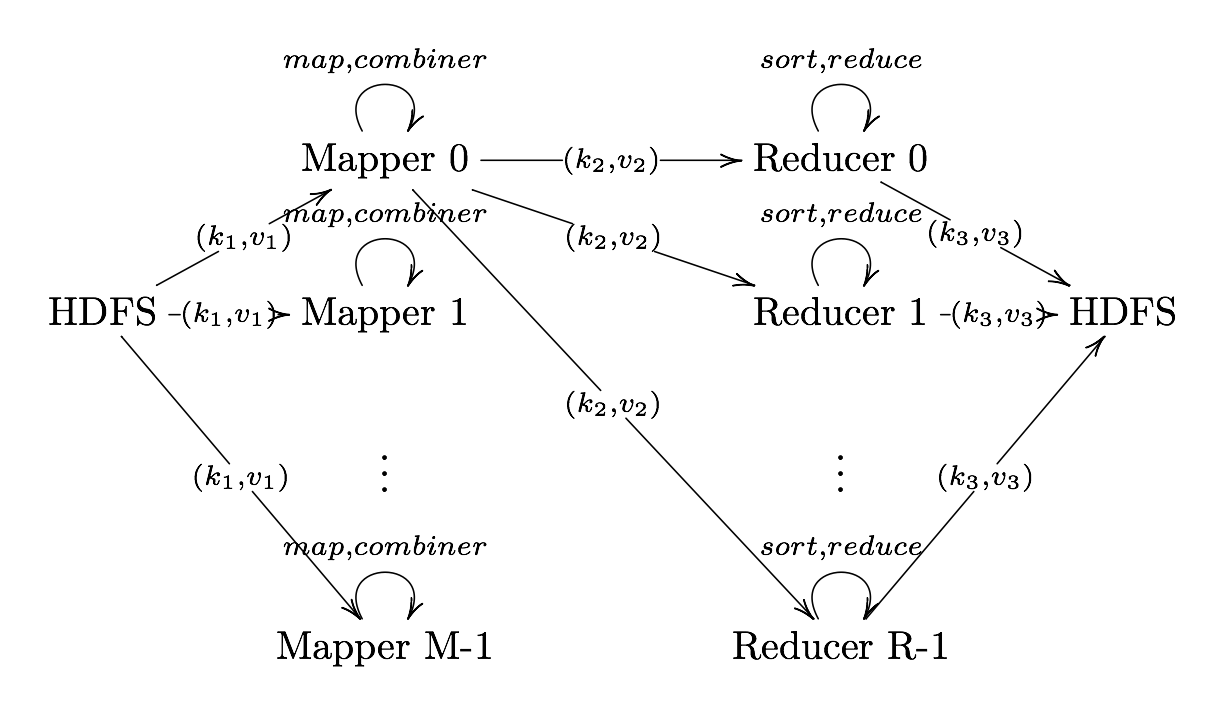
MapReduce Example: Word Count

Image Source: Xiaochong Zhang’s Blog
DAG Models
A more flexible form of MapReduce is used by Spark using Directed Acyclic Graphs (DAG).
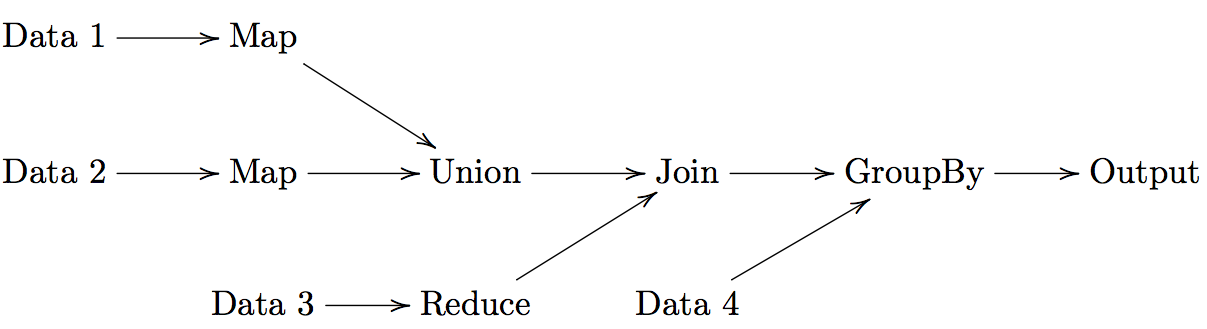
For a set of operations:
- Create a DAG for operations
- Divide DAG into tasks
- Assign tasks to nodes
MapReduce Programming Models
- Looking for parameter(s) () of a model (mean, parameters of regression, etc.)
- Partition and Model:
- Partition data,
- Apply unbiased estimator,
- Average results.
- Sketching / Sufficient Statistics:
- Partition data,
- Reduce dimensionality of data applicable to model (sufficient statistic or sketch),
- Construct model from sufficient statistic / sketch.
Partition and Model
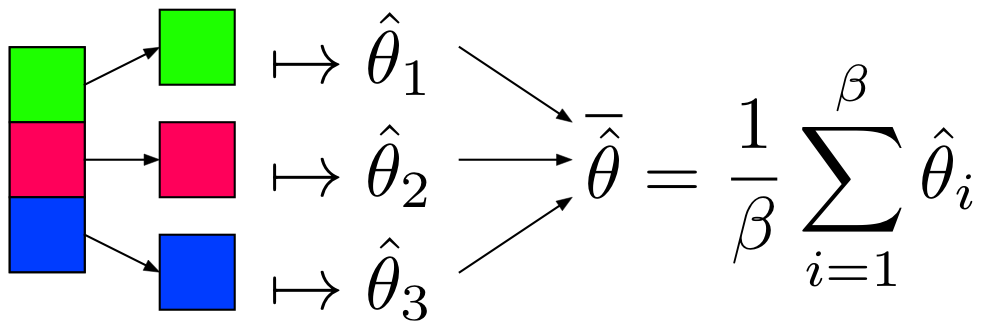
Notes
is as efficient as with the whole dataset when: 1. 2. x is IID and 3. x is equally partitioned
- Can use algorithms already in R, Python to get
- by Central Limit Theorem
Restrictions
- Values must be IID (i.e. not sorted, etc.)
- Model must produce an unbiased estimator of , denoted
Sketching / Sufficient Statistics
Streaming (Online) Algorithm: An algorithm in which the input is processed item by item. Due to limited memory and processing time, the algorithm produces a summary or “sketch” of the data.
Sufficient Statistic: A statistic with respect to a model and parameter , such that no other statistic from the sample will provide additional information.
- Sketching / Sufficient Statistics programming model aims to break down the algorithm into sketches which is then passed off into the next phase of the algorithm.
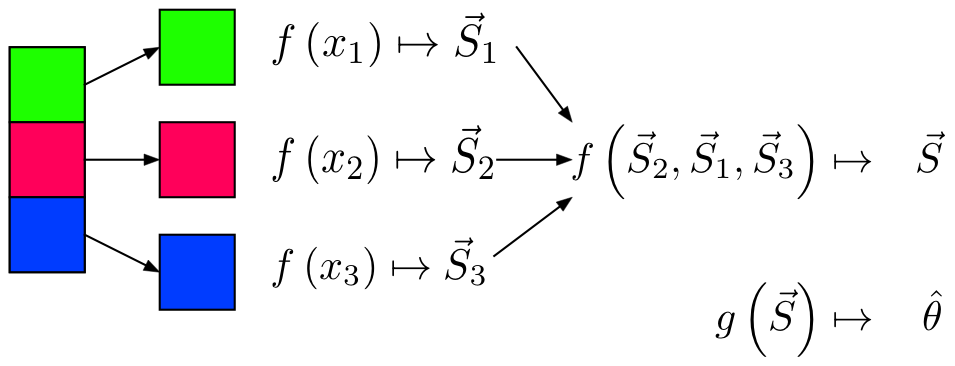
Notes
- Not all algorithms can be broken down this way.
Restrictions
- All sketches must be communicative and associative
- For each item to be processed, the order of operations must not matter
Example: Mean
Partition and Model
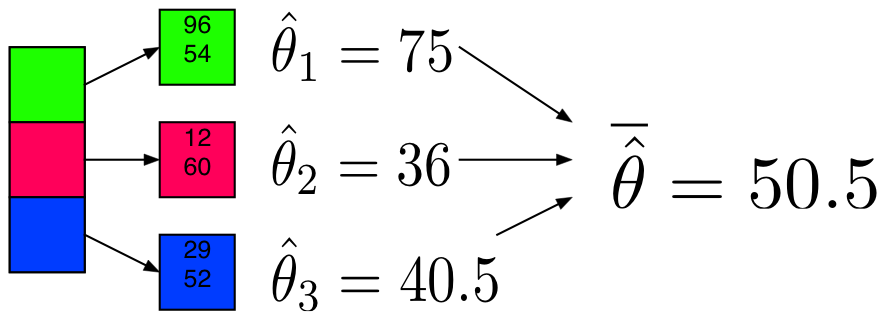
Sufficient Statistic / Sketch
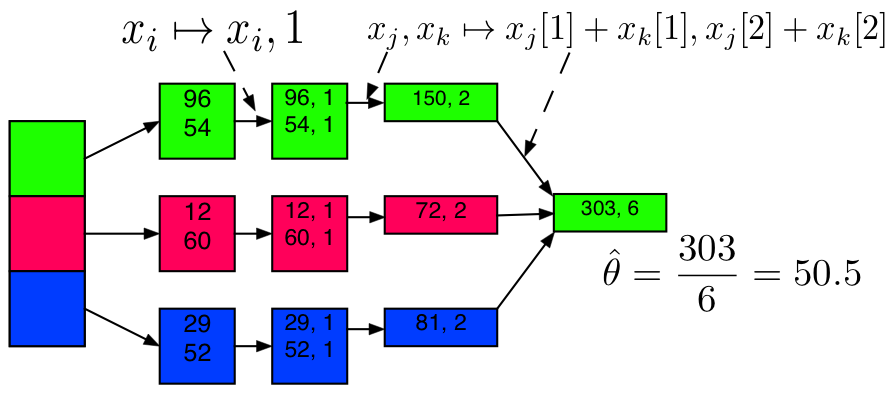
Example: Mean Code
from numpy.random import randint, chisquare
import numpy as np
X = sc.parallelize(randint(0, 101, 200000))
X_part = X.repartition(10).cache()
Example: Mean + Partition and Model
def mean_part(interator):
x = 0
n = 0
for i in interator:
x+=i
n+=1
avg = x / float(n)
return [[avg, n]]
model_mean_part = X_part.mapPartitions(mean_part) \
.collect()
model_mean_part
[[50.336245888157897, 19456],
[50.215136718750003, 20480],
[50.007421874999999, 20480],
[50.135214401294498, 19776],
[50.141858552631582, 19456],
[50.08115748355263, 19456],
[50.2578125, 20480],
[50.243945312500003, 20480],
[49.786543996710527, 19456],
[50.072363281249999, 20480]]Example: Mean + Partition and Model
The partitions are not equally sized, thus we’ll use a weighted average.
def (theta):
total = 0
weighted_avg = 0
for i in xrange(len(theta)):
weighted_avg += theta[i][0] * theta[i][1]
total+= theta[i][1]
theta_bar = weighted_avg / total
return theta_bar
print("Mean via Partition and Model:")
weighted_avg(model_mean_part)
Mean via Partition and Model:
50.128590000000003Example: Mean + Sufficient Statistics / Sketching
sketch_mean = X_part.map(lambda num: (num, 1)) \
.reduce(lambda x, y: (x[0]+y[0], x[1]+y[1]) )
x_bar_sketch = sketch_mean[0] / float(sketch_mean[1])
print("Mean via Sketching:")
x_bar_sketch
Mean via Sketching:
50.128590000000003Example: Variance
Example: Variance + Partition and Model
def var_part(interator):
x = 0
x2 = 0
n = 0
for i in interator:
x += i
x2 += i **2
n += 1
avg = x / float(n)
var = (x2 + n * avg ** 2) / (n-1)
return [[var, n]]
var_part_model = X_part.mapPartitions(var_part) \
.collect()
print("Variance via Partitioning:")
print(weighted_avg(var_part_model))
Variance via Partitioning:
851.095985421Example: Variance + Sufficient Statistics / Sketching
sketch_var = X_part.map(lambda num: (num, num**2, 1)) \
.reduce(lambda x, y: (x[0]+y[0], x[1]+y[1], x[2]+y[2]) )
x_bar_4 = sketch_var[0] / float(sketch_var[2])
N = sketch_var[2]
print("Variance via Sketching:")
(sketch_var[1] + N * x_bar_4 ** 2 ) / (N -1)
Variance via Sketching:
851.07917000774989Linear Regression
To understand the MapReduce framework, lets solve a familar problem of Linear Regression. For Hadoop/MapReduce to work we MUST figure out how to parallelize our code, in other words how to use the hadoop system to only need to make a subset of our calculations on a subset of our data.
Assumption: The value of p, the number of explanatory variables is small enough for R to easily handle i.e.
We know from linear regression, that our estimate of :
and is small enough for R to solve for , thus we only need to get .
To break up this calculation we break our matrix X into submatricies :
Sys.setenv("HADOOP_PREFIX"="/usr/local/hadoop/2.6.0")
Sys.setenv("HADOOP_CMD"="/usr/local/hadoop/2.6.0/bin/hadoop")
Sys.setenv("HADOOP_STREAMING"=
"/usr/local/hadoop/2.6.0/share/hadoop/tools/lib/hadoop-streaming-2.6.0.jar")
library(rmr2)
library(data.table)
# Setup variables
p = 10
num.obs = 200
beta.true = 1:(p+1)
X = cbind(rep(1,num.obs), matrix(rnorm(num.obs * p),
ncol = p))
y = X %*% beta.true + rnorm(num.obs)
X.index = to.dfs(cbind(y, X))
rm(X, y, num.obs, p) map.XtX = function(., Xi) {
Xi = Xi[,-1] #Get rid of y values in Xi
keyval(1, list(t(Xi) %*% Xi))
}
map.Xty = function(., Xi) {
yi = Xi[,1] # Retrieve the y values
Xi = Xi[,-1] #Get rid of y values in Xi
keyval(1, list(t(Xi) %*% yi))
}
Sum = function(., YY) {
keyval(1, list(Reduce('+', YY)))
}
Sys.setenv("HADOOP_PREFIX"="/usr/local/hadoop/2.6.0")
Sys.setenv("HADOOP_CMD"="/usr/local/hadoop/2.6.0/bin/hadoop")
Sys.setenv("HADOOP_STREAMING"=
"/usr/local/hadoop/2.6.0/share/hadoop/tools/lib/hadoop-streaming-2.6.0.jar")
library(rmr2)
library(data.table)
#Setup variables
p = 10
num.obs = 200
beta.true = 1:(p+1)
X = cbind(rep(1,num.obs), matrix(rnorm(num.obs * p),
ncol = p))
y = X %*% beta.true + rnorm(num.obs)
X.index = to.dfs(cbind(y, X))
rm(X, y, num.obs, p)
##########################
map.XtX = function(., Xi) {
Xi = Xi[,-1] #Get rid of y values in Xi
keyval(1, list(t(Xi) %*% Xi))
}
map.Xty = function(., Xi) {
yi = Xi[,1] # Retrieve the y values
Xi = Xi[,-1] #Get rid of y values in Xi
keyval(1, list(t(Xi) %*% yi))
}
Sum = function(., YY) {
keyval(1, list(Reduce('+', YY)))
}The reason we are returning a list in the map function is because otherwise Reduce will only return a some of the elements of the matricies.
List prevents this by Reduce iterating though the elements of the list (the individual matricies) and applying the binary function ‘+’ to each one.
List is used in the reduce function Sum because we will also use this as a combiner function and if we didn’t use a list we would have the same problem as above.
XtX = values(from.dfs(
mapreduce(input = X.index,
map = map.XtX,
reduce = Sum,
combine = TRUE)))[[1]]
Xty = values(from.dfs(
mapreduce(
input = X.index,
map = map.Xty,
reduce = Sum,
combine = TRUE)))[[1]]
beta.hat = solve(XtX, Xty)
print(beta.hat)
XtX = values(from.dfs(
mapreduce(input = X.index,
map = map.XtX,
reduce = Sum,
combine = TRUE)))[[1]]
Xty = values(from.dfs(
mapreduce(
input = X.index,
map = map.Xty,
reduce = Sum,
combine = TRUE)))[[1]]
beta.hat = solve(XtX, Xty)
print(beta.hat)
[,1]
[1,] 1.045835
[2,] 1.980511
[3,] 2.993829
[4,] 4.011599
[5,] 5.074755
[6,] 6.008534
[7,] 6.947164
[8,] 8.024570
[9,] 9.024757
[10,] 9.888609
[11,] 10.893023